趋化因子在高致病性人类冠状病毒感染病程中的意义
作者:吴 宪 李永哲
【摘要】目前,对SARS-CoV-2感染后宿主免疫应答已广泛研究,细胞因子风暴与COVID-19患者的疾病进展密切相关,尤其是重症或危重症患者。SARS-CoV-2,SARS-CoV和MERS-CoV在系统发育与临床感染特征具有相似之处。现有研究表明,“细胞因子风暴”是免疫系统活化过程中,大量细胞因子和趋化因子释放导致全身异常炎症反应,是引起重症感染甚至致死的关键诱因。尽管SARS和MERS的发病机制进行了多年的研究,但导致“细胞因子风暴”的特定宿主因素和关键调控机制仍不明确。鉴于SARS-CoV-2的流行,本文重点关注高致病性冠状病毒感染进程中关键趋化因子的变化情况与疾病严重程度的关系。调控趋化因子的分泌以及信号路径可能为疾病治疗提供新靶点,以缓解疾病进展,降低死亡率。
【关键词】高致病性人类冠状病毒,细胞因子风暴,趋化因子,疾病进程
趋化因子作为细胞因子超家族成员与G蛋白偶联的趋化因子受体(GPCRs)结合,调节免疫细胞的迁移和定位,进而发挥生物学功能[1, 2]。GPCRs将细胞外信号传递至下游效应器并激活多种信号路径,在不同的生理和病理条件下调节炎症反应,细胞存活,增殖和迁移[3-5]。趋化因子可诱导淋巴细胞分化为高炎症表型,抑制Treg细胞的发育,免疫细胞过度浸润,血管通透性增加[6-10]。持续异常的系统性炎症反应和过量的趋化因子的产生可导致急性肺损伤(ALI)和急性呼吸窘迫综合征(ARDS)等一系列严重的病理表现,随后可能出现淋巴细胞减少、中性粒细胞减少,弥散性血管内凝血和多器官衰竭[10-13]。目前已有大量研究报道高致病性人类冠状病毒(hCoVs)感染后趋化因子的分泌情况并且与疾病严重程度密切相关。通过总结SARS-CoV-2感染者与严重急性呼吸综合征(SARS-CoV)和中东呼吸综合征(MERS-CoV)感染进程中趋化因子表达情况的异同,深入了解诱导趋化因子释放的免疫介导事件将有助于预测预后和靶向治疗策略的研发。
一、高致病性冠状病毒感染者趋化因子的变化
1. SARS-CoV:SARS-CoV结合ACE2受体感染呼吸道和肺泡上皮细胞,树突状细胞(DC)和单核/巨噬细胞等,释放大量趋化因子。感染早期CCL2、CXCL8、CXCL9和CXCL10在外周血中升高[8, 14-16],且 CCL2,CXCL9,CXCL10在死亡组明显高于生存组[15]。其中CXCL10已被证明是早期SARS-CoV感染的独立预后因素,其上调预示着预后不良[17]。此外,CCL2、CCL5、CXCL8和CXCL10在疾病发展过程中均高于正常水平[16, 18]。与未感染者相比,SARS感染者肺泡灌洗液(BALF)中CCL2、CCL5、CXCL8和CXCL9的水平持续升高[19]。咽喉冲洗标本中也发现CXCL8呈现延迟升高的趋势[20]。
2. MERS-CoV:在MERS-CoV感染者血清/血浆中CCL2,CCL3,CCL5,CCL11,CCL22,CXCL1,CXCL8,CXCL10水平高于未感染者[21]。CCL2和CXCL10在MERS-CoV感染急性期显著升高,恢复期下降至基线水平,提示两者与疾病发展进程以及严重程度有关[22, 23]。
3. SARS-CoV-2:SARS-CoV-2感染者血浆/血清中的多种趋化因子显著升高,包括CCL2、CCL3、CCL5、CCL8、CCL11、CCL20、CCL27、CXCL2、CXCL8、CXCL9、CXCL10、CXCL13和CXCL16[24-31],其中,CXCL8、CXCL10、CXCL13与疾病严重程度相关[26, 27]。CCL2在感染早期升高,发病后2周左右即可恢复至正常水平[30]。轻度感染者CCL5在感染早期即开始显著升高[32],但在严重SARS-CoV-2感染CCL5下降并且与疾病严重性呈负相关[26]。
为了清晰、直观地观察趋化因子的动态变化,一些研究依据COVID-19感染者的临床表现分为无症状组、轻/中度组、重度组和危重组,多时间点采集不同严重程度患者的血浆/血清标本进行检测,进而深入探索趋化因子与疾病严重性的关系。除上述分泌升高的趋化因子外,在感染者血浆/血清发现CLL4、CCL7、CXCL1和CXCL12也呈现明显增加趋势[31, 33-35]。高水平CCL2,CCL3,CCL4,CCL5,CCL7,CXCL9与疾病的严重性密切相关[32-35]。其中CCL3,CCL7,CXCL9,CXCL10在危重症感染者中高于重度和中度感染者[33, 35],在感染晚期,CCL7、CXCL9和CXCL10并未进一步上调甚至降低,表明这些趋化因子与疾病的严重程度相关[35]。与未感染组相比,CXCL10在无症状感染者组即出现升高,在有症状组呈显著升高,而在恢复期CXCL10水平恢复正常。另外,轻/中度感染者血清中CXCL8、CCL11、CXCL1、CXCL10和CXCL9水平升高,而重度感染者血清CCL2、CCL3和CCL7水平升高[34]。
为明确重症肺炎的预测标志物,有研究比较重症感染者组和轻症组的趋化因子分泌情况。趋化因子CCL2、CCL5、CXCL8、CXCL9、CXCL10在重度/危重COVID-19患者中显著高于非重度患者,CCL2、CCL17、CXCL8、CXCL9、CXCL10可预测严重/危重症状的发生[36-41]。值得注意的是,CCL17在SARS-CoV-2感染早期,可区分轻度/中度组和重度/严重组[37]。此外有研究表明,与非重症监护病房(ICU)患者相比,ICU患者的CCL2、CCL3、CCL7、CCL8、CXCL8、CXCL10和CXCL11明显增加[31, 42, 43]。同时,ICU患者CCL5明显低于非ICU患者,CCL7和CXCL8的联合应用对临床的潜在风险分级具有重要意义[43]。另外,与存活组相比,死亡组患者CXCL8水平显著持续升高并且与死亡率呈正相关[44, 45]。
除检测血清/血浆样本中趋化因子外,在SARS-CoV-2感染者鼻咽拭子样本检测到的趋化因子与对照组相比,CXCL8,CXCL9,CXCL10明显上调[46],而CCL5表达低于对照组[47]。ICU患者支气管肺泡灌洗液(BALF)中CXCL8明显高于非ICU患者[48]。此外,通过对鼻咽和支气管样本进行单细胞RNA测序发现,重症感染者较中度感染者CCL2、CCL3、CCL20、CXCL1、CXCL3、CXCL8和CXCL10表达升高[49]。支气管肺泡液BALF和PBMC标本的转录组分析结果表明,患者的趋化因子水平,包括BALF中的CXCL1、CXCL2、CXCL6、CCL8、CXCL10、CCL2、CCL3、CCL4和PBMC中的CXCL10,均显著高于健康对照组[50]。
根据以上总结,趋化因子在疾病进程中呈动态变化且受多种因素影响,深入探索趋化因子变化规律及调控机制,才能为准确判断疾病的严重性以及感染预后提供重要依据。
二、 趋化因子异常分泌的潜在机制
1. IL-6驱动的免疫失调:高致病性hCoVs感染导致多种类型的免疫细胞活化,包括单核细胞、巨噬细胞和中性粒细胞,并伴有IL-6的分泌增加。持续的IL-6产生触发级联反应,引起大量细胞因子释放,最终导致冠状病毒感染的免疫病理变化,如:急性呼吸窘迫综合征[51, 52]。IL-6经经典的顺式或反式途径可激活多种免疫细胞,在顺式信号转导中,IL-6结合IL-6膜受体(mIL-6R),信号转导涉及JAKs和STAT3通路[53]。在反式信号转导中,体液高浓度的IL-6与可溶性IL-6R(sIL-6R)结合,与gp130二聚体形成复合物,触发IL-6/sIL-6R/JAK/STAT3信号传导,激活多种不表达mIL-6R的细胞,如内皮细胞。活化的细胞分泌大量的促炎性细胞因子和趋化因子,可能导致CCL2和CXCL8分泌的全身性“细胞因子风暴”[54](图1)。
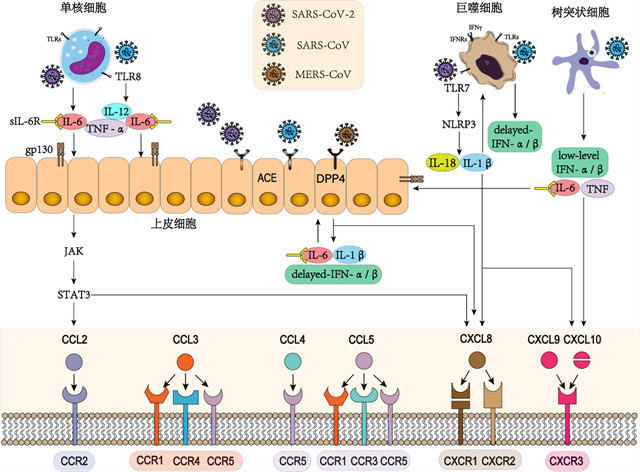
(1)IL-6驱动的免疫失调:IL-6结合sIL-6R,与gp130二聚体形成复合物,激活内皮细胞IL-6/sIL-6R/JAK/STAT3信号传导。活化的细胞分泌促炎细胞因子和趋化因子,包括CCL2和CXCL8。SARS-CoV可通过激活单核细胞TLR8诱导TNF-α,IL-6和IL-12的释放。(2)IL-1β诱导巨噬细胞异常活化:巨噬细胞TLR7活化刺激NLRP3炎症小体,导致成熟IL-1β和/或IL-18的释放。IL-1β增强单核细胞源性巨噬细胞的活化,引起CXCL8和CXCL10释放。(3)IFN应答失衡:SARS-CoV感染的巨噬细胞诱导IFN和其他促炎细胞因子的延迟分泌。此外,SARS-CoV感染DCs可导致I型干扰素(IFN-α/β)的下调和TNF,IL-6的中度升高。而MERS-CoV感染在内皮细胞中可诱导释放IFN-α/β,IL-1β,IL-6,CXCL8。
图1. hCoVs感染过程中趋化因子调控机制示意图
2. IL-1β诱导的巨噬细胞异常活化:IL-1β是另一种参与多种细胞因子风暴综合征的促炎细胞因子。病毒通过ACEs进入细胞质并激活NLRP3炎性体,通过TLR7介导的Ⅰ型干扰素反应产生IL-1β,进而以自分泌或旁分泌方式放大单核细胞源性巨噬细胞的激活,从而促进多种免疫细胞分泌促炎性因子,包括CXCL8和CXCL10,而导致“细胞因子风暴”[55, 56]。由促炎细胞因子TNFα和IL-1β快速诱导CXCL8释放,激活PI3K/MAPK信号和Raf-1/MAP/ERK信号级联反应[57, 58]。 SARS-CoV感染也可以调控NLRP3炎症体依赖CXCL10介导的肺部炎症[59]。SARS-CoV的蛋白(S蛋白和非结构蛋白1)可激活外周血单个核细胞(PBMC),单核细胞系(THP-1)以及肺上皮细胞通过激活NF-κB迅速表达CCL3、CCL5和CXCL10[60, 61]。若利用重组人IL-1受体拮抗剂阻断IL-1β可显著改善急性细胞因子风暴综合征患者的预后,证实IL-1β在冠状病毒诱导的过度炎症应答反应中的重要作用(图1)。
3. 干扰素应答不平衡:在感染的早期和急性重症阶段,异常的干扰素(IFN)应答会加速病毒血症的进展,而SARS患者过度炎性应答和趋化因子的释放可能导致免疫细胞大量浸润和免疫功能失调[56]。研究表明SARS-CoV可调节并阻止感染细胞诱导抗病毒I型干扰素分泌[56, 62]。另外,SARS-CoV感染树突状细胞(DCs)可导致I型干扰素(IFN-α/β)的下调,促炎细胞因子TNF和IL-6升高,炎症趋化因子的合成和分泌显著增加。此外,SARS-CoV感染的巨噬细胞延迟诱导IFN和其他促炎细胞因子释放[6, 7, 10]。同样,MERS-CoV感染可诱导人呼吸道内皮细胞显著但延迟释放IFN-α/β和促炎细胞因子IL-1β、IL-6和CXCL8 [63]。抗病毒细胞因子IFN-α/β的异常分泌导致病毒快速复制,刺激内皮细胞、上皮细胞和天然免疫细胞分泌大量促炎细胞因子和趋化因子。促炎性趋化因子介导单核细胞/巨噬细胞早期进入肺部,产生刺激/分泌的恶性循环,导致细胞因子风暴以及致命的ALI和ARDS(图1)。
三、结论
本综述旨在总结高致病性hCoVs感染过程中趋化因子的分泌情况以及调控机制的研究进展。趋化因子系统在机体免疫应答的启动、激活和调节过程中起到重要作用。因此,了解趋化因子在疾病进程中的分泌情况以及异常释放机制,对于制定有效的治疗策略缓解疾病进展以及降低高致病性hCoVs感染相关的死亡率至关重要。
参考文献
Zlotnik, A.,O. Yoshie. Chemokines: a new classification system and their role in immunity[J]. Immunity, 2000, 12(2): 121-7.
Wang, Z., H. Shang,Y. Jiang. Chemokines and Chemokine Receptors: Accomplices for Human Immunodeficiency Virus Infection and Latency[J]. Front Immunol, 2017, 8: 1274.
Stone, M.J., J.A. Hayward, C. Huang, et al. Mechanisms of Regulation of the Chemokine-Receptor Network[J]. Int J Mol Sci, 2017, 18(2).
Nibbs, R.J.,G.J. Graham. Immune regulation by atypical chemokine receptors[J]. Nat Rev Immunol, 2013, 13(11): 815-29.
Suresh, P.,A. Wanchu. Chemokines and chemokine receptors in HIV infection: role in pathogenesis and therapeutics[J]. J Postgrad Med, 2006, 52(3): 210-7.
Cheung, C.Y., L.L. Poon, I.H. Ng, et al. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis[J]. J Virol, 2005, 79(12): 7819-26.
Law, H.K., C.Y. Cheung, H.Y. Ng, et al. Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells[J]. Blood, 2005, 106(7): 2366-74.
Chien, J.Y., P.R. Hsueh, W.C. Cheng, et al. Temporal changes in cytokine/chemokine profiles and pulmonary involvement in severe acute respiratory syndrome[J]. Respirology, 2006, 11(6): 715-22.
Kuypers, J., E.T. Martin, J. Heugel, et al. Clinical disease in children associated with newly described coronavirus subtypes[J]. Pediatrics, 2007, 119(1): e70-6.
Channappanavar, R.,S. Perlman. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology[J]. Semin Immunopathol, 2017, 39(5): 529-539.
Chen, G., D. Wu, W. Guo, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019[J]. J Clin Invest, 2020, 130(5): 2620-2629.
Huang, C., Y. Wang, X. Li, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China[J]. Lancet, 2020.
Cameron, M.J., J.F. Bermejo-Martin, A. Danesh, et al. Human immunopathogenesis of severe acute respiratory syndrome (SARS) [J]. Virus Res, 2008, 133(1): 13-9.
Jiang, Y., J. Xu, C. Zhou, et al. Characterization of cytokine/chemokine profiles of severe acute respiratory syndrome[J]. Am J Respir Crit Care Med, 2005, 171(8): 850-7.
Huang, K.J., I.J. Su, M. Theron, et al. An interferon-gamma-related cytokine storm in SARS patients[J]. J Med Virol, 2005, 75(2): 185-94.
Wong, C.K., C.W. Lam, A.K. Wu, et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome[J]. Clin Exp Immunol, 2004, 136(1): 95-103.
Tang, N.L., P.K. Chan, C.K. Wong, et al. Early enhanced expression of interferon-inducible protein-10 (CXCL-10) and other chemokines predicts adverse outcome in severe acute respiratory syndrome[J]. Clin Chem, 2005, 51(12): 2333-40.
Ward, S.E., M.R. Loutfy, L.M. Blatt, et al. Dynamic changes in clinical features and cytokine/chemokine responses in SARS patients treated with interferon alfacon-1 plus corticosteroids[J]. Antivir Ther, 2005, 10(2): 263-75.
Wang, C.H., C.Y. Liu, Y.L. Wan, et al. Persistence of lung inflammation and lung cytokines with high-resolution CT abnormalities during recovery from SARS[J]. Respir Res, 2005, 6(1): 42.
Wang, W.K., S.Y. Chen, I.J. Liu, et al. Temporal relationship of viral load, ribavirin, interleukin (IL)-6, IL-8, and clinical progression in patients with severe acute respiratory syndrome[J]. Clin Infect Dis, 2004, 39(7): 1071-5.
Min, C.K., S. Cheon, N.Y. Ha, et al. Comparative and kinetic analysis of viral shedding and immunological responses in MERS patients representing a broad spectrum of disease severity[J]. Sci Rep, 2016, 6: 25359.
Kim, E.S.,P.G. Choe. Clinical Progression and Cytokine Profiles of Middle East Respiratory Syndrome Coronavirus Infection[J], 2016, 31(11): 1717-1725.
Shin, H.S., Y. Kim, G. Kim, et al. Immune Responses to Middle East Respiratory Syndrome Coronavirus During the Acute and Convalescent Phases of Human Infection[J]. Clin Infect Dis, 2019, 68(6): 984-992.
Gibellini, L.,S. De Biasi. Altered bioenergetics and mitochondrial dysfunction of monocytes in patients with COVID-19 pneumonia[J], 2020, 12(12): e13001.
Del Valle, D.M., S. Kim-Schulze, H.H. Huang, et al. An inflammatory cytokine signature predicts COVID-19 severity and survival[J], 2020, 26(10): 1636-1643.
Horspool, A.M., T. Kieffer, B.P. Russ, et al. Interplay of Antibody and Cytokine Production Reveals CXCL13 as a Potential Novel Biomarker of Lethal SARS-CoV-2 Infection[J], 2021, 6(1).
Hue, S., A. Beldi-Ferchiou, I. Bendib, et al. Uncontrolled Innate and Impaired Adaptive Immune Responses in Patients with COVID-19 Acute Respiratory Distress Syndrome[J], 2020, 202(11): 1509-1519.
Blanco-Melo, D., B.E. Nilsson-Payant, W.C. Liu, et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19[J]. Cell, 2020, 181(5): 1036-1045.e9.
Hou, X., X. Zhang, X. Wu, et al. Serum Protein Profiling Reveals a Landscape of Inflammation and Immune Signaling in Early-stage COVID-19 Infection[J], 2020, 19(11): 1749-1759.
Young, B.E., S.W.X. Ong, L.F.P. Ng, et al. Viral dynamics and immune correlates of COVID-19 disease severity[J]. Clin Infect Dis, 2020.
Huang, C., Y. Wang, X. Li, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China[J]. Lancet, 2020, 395(10223): 497-506.
Zhao, Y., L. Qin, P. Zhang, et al. Longitudinal COVID-19 profiling associates IL-1RA and IL-10 with disease severity and RANTES with mild disease[J]. JCI Insight, 2020, 5(13).
Yang, Y., C. Shen, J. Li, et al. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19[J]. J Allergy Clin Immunol, 2020, 146(1): 119-127.e4.
Chi, Y., Y. Ge, B. Wu, et al. Serum Cytokine and Chemokine Profile in Relation to the Severity of Coronavirus Disease 2019 in China[J]. J Infect Dis, 2020, 222(5): 746-754.
Xu, Z.S., T. Shu, L. Kang, et al. Temporal profiling of plasma cytokines, chemokines and growth factors from mild, severe and fatal COVID-19 patients[J]. Signal Transduct Target Ther, 2020, 5(1): 100.
Li, S., L. Jiang, X. Li, et al. Clinical and pathological investigation of patients with severe COVID-19[J]. JCI Insight, 2020, 5(12).
Sugiyama, M., N. Kinoshita, S. Ide, et al. Serum CCL17 level becomes a predictive marker to distinguish between mild/moderate and severe/critical disease in patients with COVID-19[J]. Gene, 2021, 766: 145145.
Tincati, C., E.S. Cannizzo, M. Giacomelli, et al. Heightened Circulating Interferon-Inducible Chemokines, and Activated Pro-Cytolytic Th1-Cell Phenotype Features Covid-19 Aggravation in the Second Week of Illness[J]. Front Immunol, 2020, 11: 580987.
Chen, Y., J. Wang, C. Liu, et al. IP-10 and MCP-1 as biomarkers associated with disease severity of COVID-19[J], 2020, 26(1): 97.
Kwon, J.S., J.Y. Kim, M.C. Kim, et al. Factors of Severity in Patients with COVID-19: Cytokine/Chemokine Concentrations, Viral Load, and Antibody Responses[J]. Am J Trop Med Hyg, 2020, 103(6): 2412-2418.
Zhang, X.,Y. Tan. Viral and host factors related to the clinical outcome of COVID-19[J], 2020, 583(7816): 437-440.
Haljasmägi, L., A. Salumets,A.P. Rumm. Longitudinal proteomic profiling reveals increased early inflammation and sustained apoptosis proteins in severe COVID-19[J], 2020, 10(1): 20533.
Chen, L., G. Wang, J. Tan, et al. Scoring cytokine storm by the levels of MCP-3 and IL-8 accurately distinguished COVID-19 patients with high mortality[J]. Signal Transduct Target Ther, 2020, 5(1): 292.
[44] Li, Q., W. Xu, W.X. Li, et al. Dynamics of cytokines and lymphocyte subsets associated with the poor prognosis of severe COVID-19[J]. Eur Rev Med Pharmacol Sci, 2020, 24(23): 12536-12544.
Balnis, J., A.P. Adam, A. Chopra, et al. Unique inflammatory profile is associated with higher SARS-CoV-2 acute respiratory distress syndrome (ARDS) mortality[J]. Am J Physiol Regul Integr Comp Physiol, 2021.
Lieberman, N.A.P.,V. Peddu. In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age[J], 2020, 18(9): e3000849.
Montalvo Villalba, M.C., O. Valdés Ramírez, M. Muné Jiménez, et al. Interferon gamma, TGF-β1 and RANTES expression in upper airway samples from SARS-CoV-2 infected patients[J]. Clin Immunol, 2020, 220: 108576.
Pandolfi, L., T. Fossali, V. Frangipane, et al. Broncho-alveolar inflammation in COVID-19 patients: a correlation with clinical outcome[J]. BMC Pulm Med, 2020, 20(1): 301.
Chua, R.L.,S. Lukassen. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis[J], 2020, 38(8): 970-979.
Xiong, Y., Y. Liu, L. Cao, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients[J], 2020, 9(1): 761-770.
Giamarellos-Bourboulis, E.J., M.G. Netea, N. Rovina, et al. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure[J]. Cell Host Microbe, 2020, 27(6): 992-1000.e3.
Patterson, B.K., H. Seethamraju, K. Dhody, et al. Disruption of the CCL5/RANTES-CCR5 Pathway Restores Immune Homeostasis and Reduces Plasma Viral Load in Critical COVID-19[J], 2020.
Kang, S., T. Tanaka, M. Narazaki, et al. Targeting Interleukin-6 Signaling in Clinic[J]. Immunity, 2019, 50(4): 1007-1023.
Moore, J.B.,C.H. June. Cytokine release syndrome in severe COVID-19[J]. Science, 2020, 368(6490): 473-474.
Merad, M.,J.C. Martin. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages[J], 2020, 20(6): 355-362.
Thiel, V.,F. Weber. Interferon and cytokine responses to SARS-coronavirus infection[J]. Cytokine Growth Factor Rev, 2008, 19(2): 121-32.
Hoffmann, E., O. Dittrich-Breiholz, H. Holtmann, et al. Multiple control of interleukin-8 gene expression[J]. J Leukoc Biol, 2002, 72(5): 847-55.
Knall, C., S. Young, J.A. Nick, et al. Interleukin-8 regulation of the Ras/Raf/mitogen-activated protein kinase pathway in human neutrophils[J]. J Biol Chem, 1996, 271(5): 2832-8.
Chang, Y.S., B.H. Ko, J.C. Ju, et al. SARS Unique Domain (SUD) of Severe Acute Respiratory Syndrome Coronavirus Induces NLRP3 Inflammasome-Dependent CXCL10-Mediated Pulmonary Inflammation[J], 2020, 21(9).
Dosch, S.F., S.D. Mahajan,A.R. Collins. SARS coronavirus spike protein-induced innate immune response occurs via activation of the NF-kappaB pathway in human monocyte macrophages in vitro[J]. Virus Res, 2009, 142(1-2): 19-27.
Law, A.H., D.C. Lee, B.K. Cheung, et al. Role for nonstructural protein 1 of severe acute respiratory syndrome coronavirus in chemokine dysregulation[J]. J Virol, 2007, 81(1): 416-22.
Chen, J.,K. Subbarao. The Immunobiology of SARS*[J]. Annu Rev Immunol, 2007, 25: 443-72.
Chu, H., J. Zhou, B.H. Wong, et al. Middle East Respiratory Syndrome Coronavirus Efficiently Infects Human Primary T Lymphocytes and Activates the Extrinsic and Intrinsic Apoptosis Pathways[J]. J Infect Dis, 2016, 213(6): 904-14.





